Analysis
First, we begin by exporting the data. One can simply download the training and testing datasets using:
# Download the training and testing datasets download.file("https://d396qusza40orc.cloudfront.net/predmachlearn/pml-training.csv","training.csv",method = "curl") download.file("https://d396qusza40orc.cloudfront.net/predmachlearn/pml-testing.csv","testing.csv",method = "curl")
Then, we load some useful packages using:
# Require the necessary packages require(data.table) require(dplyr) require(caret)
Now let's load the data into memory:
# Load the training and testing datasets training <- tbl_df(fread("training.csv",na.strings=c('#DIV/0!', '', 'NA'))) testing <- tbl_df(fread("testing.csv",na.strings=c('#DIV/0!', '', 'NA')))
Now that we have the data in the memory, let's get to the fun part. First thing we better do is to split the training data into two parts. We'll use 70% of this data to actually train our model and the remaining 30% to validate it:
# Now split the training into to as actual testing and validation set.seed(1234) # Don't forget the reproducibility! trainingDS <- createDataPartition( y = training$classe, p = 0.7, list = FALSE) actual.training <- training[trainingDS,] actual.validation <- training[-trainingDS,]
Next, we need to prepare the data for modeling. If you look at the training data you'll see that there are a number of variables that have either no variance or a large fraction of missing values. These will not really help us in any meaningful way. Therefore, let's clean them up for a healthy modeling:
# Now clean-up the variables w/ zero variance # Be careful, kick out the same variables in both cases nzv <- nearZeroVar(actual.training) actual.training <- actual.training[,-nzv] actual.validation <- actual.validation[,-nzv] # Remove variables that are mostly NA mostlyNA <- sapply(actual.training,function(x) mean(is.na(x))) > 0.95 actual.training <- actual.training[,mostlyNA==FALSE] actual.validation <- actual.validation[,mostlyNA==FALSE] # At this point we're already down to 59 variables from 160 # See that the first 5 variables are identifiers that are # not probably useful for prediction so get rid of those # Dropping the total number of variables to 54 (53 for prediction) actual.training <- actual.training[,-(1:5)] actual.validation <- actual.validation[,-(1:5)]
At this point, we have healthy clean data that we can use for building models. We'll build two models: a random forest and a generalized boosted model. We'll train these in the training portion of the original training dataset and then test them in the validation portion of the original training dataset:
# Now let's build a random forest model set.seed(1234) modelRF <- train( classe ~., data = actual.training, method = "rf", trControl = trainControl(method="cv",number=3) )
# One can also build a generalized boosted model and compare its accuracy # to random forest model set.seed(1234) modelBM <- train( classe ~., data = actual.training, method = "gbm", trControl = trainControl(method="repeatedcv",number = 5,repeats = 1), verbose = FALSE)
Then let's see how well these two models perform predicting the values in the validation dataset. This can be easily accomplished by predicting the values in the validation set, and then comparing the predictions with the actual values.
# Now get the prediction in the validation portion and see how well we do prediction.validation.rf <- predict(modelRF,actual.validation) conf.matrix.rf <- confusionMatrix(prediction.validation.rf,actual.validation$classe) print(conf.matrix.rf)
## Confusion Matrix and Statistics ## ## Reference ## Prediction A B C D E ## A 1674 4 0 0 0 ## B 0 1134 4 0 0 ## C 0 1 1022 1 0 ## D 0 0 0 963 2 ## E 0 0 0 0 1080 ## ## Overall Statistics ## ## Accuracy : 0.998 ## 95% CI : (0.9964, 0.9989) ## No Information Rate : 0.2845 ## P-Value [Acc > NIR] : < 2.2e-16 ## ## Kappa : 0.9974 ## Mcnemar's Test P-Value : NA ## ## Statistics by Class: ## ## Class: A Class: B Class: C Class: D Class: E ## Sensitivity 1.0000 0.9956 0.9961 0.9990 0.9982 ## Specificity 0.9991 0.9992 0.9996 0.9996 1.0000 ## Pos Pred Value 0.9976 0.9965 0.9980 0.9979 1.0000 ## Neg Pred Value 1.0000 0.9989 0.9992 0.9998 0.9996 ## Prevalence 0.2845 0.1935 0.1743 0.1638 0.1839 ## Detection Rate 0.2845 0.1927 0.1737 0.1636 0.1835 ## Detection Prevalence 0.2851 0.1934 0.1740 0.1640 0.1835 ## Balanced Accuracy 0.9995 0.9974 0.9978 0.9993 0.9991
# Now get the prediction in the validation portion and see how well we do prediction.validation.bm <- predict(modelBM,actual.validation) conf.matrix.bm <- confusionMatrix(prediction.validation.bm,actual.validation$classe) print(conf.matrix.bm)
## Confusion Matrix and Statistics ## ## Reference ## Prediction A B C D E ## A 1673 14 0 0 0 ## B 1 1107 9 7 1 ## C 0 15 1017 11 3 ## D 0 3 0 946 9 ## E 0 0 0 0 1069 ## ## Overall Statistics ## ## Accuracy : 0.9876 ## 95% CI : (0.9844, 0.9903) ## No Information Rate : 0.2845 ## P-Value [Acc > NIR] : < 2.2e-16 ## ## Kappa : 0.9843 ## Mcnemar's Test P-Value : NA ## ## Statistics by Class: ## ## Class: A Class: B Class: C Class: D Class: E ## Sensitivity 0.9994 0.9719 0.9912 0.9813 0.9880 ## Specificity 0.9967 0.9962 0.9940 0.9976 1.0000 ## Pos Pred Value 0.9917 0.9840 0.9723 0.9875 1.0000 ## Neg Pred Value 0.9998 0.9933 0.9981 0.9963 0.9973 ## Prevalence 0.2845 0.1935 0.1743 0.1638 0.1839 ## Detection Rate 0.2843 0.1881 0.1728 0.1607 0.1816 ## Detection Prevalence 0.2867 0.1912 0.1777 0.1628 0.1816 ## Balanced Accuracy 0.9980 0.9841 0.9926 0.9894 0.9940
We can investigate our generalized boosted model a bit further to see which variables have the highest relative influence:
# Print the summary of our GBM print(summary(modelBM))
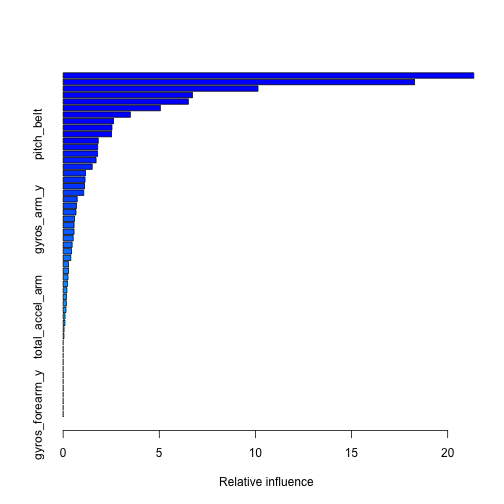
## var rel.inf ## num_window num_window 21.35545512 ## roll_belt roll_belt 18.27724938 ## pitch_forearm pitch_forearm 10.13583112 ## magnet_dumbbell_z magnet_dumbbell_z 6.72541291 ## yaw_belt yaw_belt 6.51200291 ## magnet_dumbbell_y magnet_dumbbell_y 5.05649194 ## roll_forearm roll_forearm 3.49549946 ## accel_forearm_x accel_forearm_x 2.61845321 ## magnet_belt_z magnet_belt_z 2.53921239 ## pitch_belt pitch_belt 2.51716927 ## gyros_belt_z gyros_belt_z 1.82833065 ## gyros_dumbbell_y gyros_dumbbell_y 1.79600229 ## accel_dumbbell_z accel_dumbbell_z 1.78978003 ## roll_dumbbell roll_dumbbell 1.71291149 ## accel_dumbbell_y accel_dumbbell_y 1.51172485 ## yaw_arm yaw_arm 1.16790745 ## magnet_forearm_z magnet_forearm_z 1.13838373 ## accel_forearm_z accel_forearm_z 1.11085165 ## accel_dumbbell_x accel_dumbbell_x 1.07112326 ## magnet_belt_y magnet_belt_y 0.73220505 ## roll_arm roll_arm 0.69769807 ## magnet_arm_z magnet_arm_z 0.66713578 ## gyros_arm_y gyros_arm_y 0.59235410 ## magnet_belt_x magnet_belt_x 0.56962809 ## accel_belt_z accel_belt_z 0.56689770 ## gyros_belt_y gyros_belt_y 0.52256127 ## magnet_arm_x magnet_arm_x 0.46724034 ## magnet_dumbbell_x magnet_dumbbell_x 0.43441017 ## total_accel_dumbbell total_accel_dumbbell 0.39610082 ## magnet_forearm_x magnet_forearm_x 0.28197085 ## total_accel_forearm total_accel_forearm 0.27783955 ## gyros_dumbbell_x gyros_dumbbell_x 0.25152542 ## magnet_arm_y magnet_arm_y 0.22235172 ## pitch_dumbbell pitch_dumbbell 0.19453168 ## accel_arm_z accel_arm_z 0.17252396 ## accel_forearm_y accel_forearm_y 0.16660947 ## gyros_belt_x gyros_belt_x 0.14726266 ## total_accel_arm total_accel_arm 0.09844732 ## gyros_forearm_z gyros_forearm_z 0.09281553 ## gyros_dumbbell_z gyros_dumbbell_z 0.04731855 ## magnet_forearm_y magnet_forearm_y 0.04077878 ## total_accel_belt total_accel_belt 0.00000000 ## accel_belt_x accel_belt_x 0.00000000 ## accel_belt_y accel_belt_y 0.00000000 ## pitch_arm pitch_arm 0.00000000 ## gyros_arm_x gyros_arm_x 0.00000000 ## gyros_arm_z gyros_arm_z 0.00000000 ## accel_arm_x accel_arm_x 0.00000000 ## accel_arm_y accel_arm_y 0.00000000 ## yaw_dumbbell yaw_dumbbell 0.00000000 ## yaw_forearm yaw_forearm 0.00000000 ## gyros_forearm_x gyros_forearm_x 0.00000000 ## gyros_forearm_y gyros_forearm_y 0.00000000
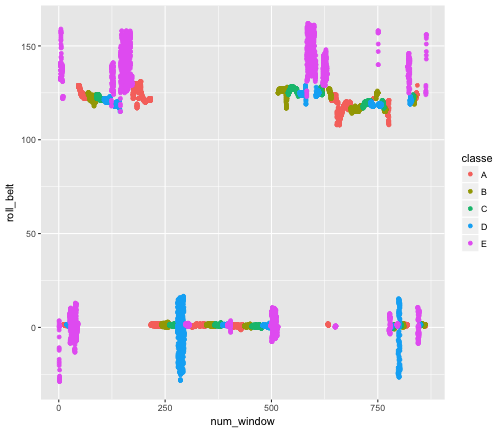
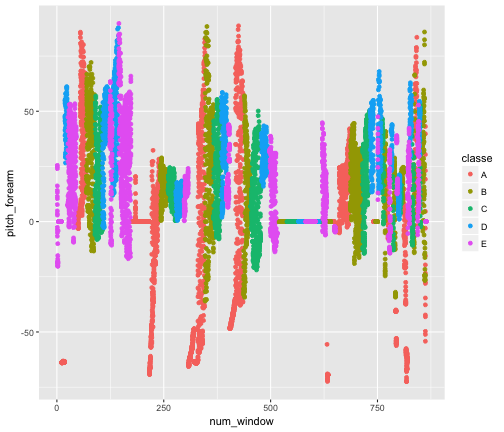
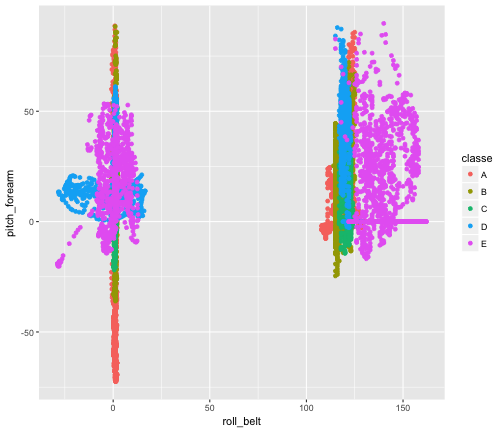
The above list shows the ranking of variables in our GBM. We see that num_window, roll_belt, and pitch_forearm are the most performant ones. We can checkout a few plots demonstrating their power:
qplot(num_window, roll_belt , data = actual.training, col = classe)
qplot(num_window, pitch_forearm, data = actual.training, col = classe)
qplot(roll_belt , pitch_forearm, data = actual.training, col = classe)
At this point we see the random forest has marginally better performance (Accuracy : 0.998) than the generalized boosted model (Accuracy : 0.9876). Actually we can go w/ either or ensemble them but that might be an overkill at this point. In any case they yield the same result. Let's test our model in the actual testing dataset:
# Now get the prediction in the testing portion and see what we get prediction.testing.rf <- predict(modelRF,testing) print(prediction.testing.rf)
## [1] B A B A A E D B A A B C B A E E A B B B ## Levels: A B C D E